imsim
Manual
The following sections briefly describe how to run IMSIM from the command-line, and details the required input and output files.
Introduction
IMSIM simulates two-dimensional small angle X-ray scattering patterns and therefore, in extension, one dimensional profiles from biological macromolecules in solution.
IMSIM implements a statistical approach yielding two-dimensional images in .tiff, .cbf or .edf format, which can be readily processed by existing radial averaging tools. The resulting intensities and error estimates of one-dimensional patterns obtained from the radial average of the two-dimensional images exhibit the same statistical properties as observed with actual experimental data. The effects due to changes of concentration, exposure time, flux, wavelength, sample-detector distance, detector dimensions, pixel size, and the mask as well as incident beam position can be considered for the simulation.
Running imsim
Usage:
$ imsim [OPTIONS] <ABSFILE>
OPTIONS known by IMSIM are described in next section, the required argument is described in the section on input files.
Command-Line Arguments and Options
IMSIM requires the following command-line arguments:
| Argument | Description |
|---|---|
| ABSFILE | Exactly one calculated SAS intensity data (.abs) file. |
Absolute as well as relative paths to files are accepted. Up to one of the input files may also be given as ‘-‘, in this case input is read from stdin instead of a file.
IMSIM recognizes the following command line options:
| Short Option | Long Option | Description |
|---|---|---|
| --seed <INT> | Set the seed for the random number generator; default: current time | |
| --background <FILE> | Calculated background intensity in I[cm^-1^] / conc[mg/ml]; default: built-in flat background | |
| --detector <NAME> | Detector name, defines pixel size and mask dimensions. One of: Pilatus100k, Pilatus300k, Pilatus1M, Pilatus2M, Pilatus6M, Eiger500k, Eiger1M, Eiger4M, Eiger9M, Eiger16M; default: Pilatus1M | |
| --detector-distance <m> | Sample-detector-distance in meter; default: 1.0 | |
| --flux <N> | Number of incoming virtual photons per second; default: 10e12 | |
| -p | --scattering-probability <P> | Probability that a virtual photons scatters; default: 1e-6 |
| --exptime <T> | Exposure time in seconds; default: 1 | |
| -c | --concentration <C> | Factor proportional to concentration of sample where a concentration of 0.0 corresponds to background simulation.; default: 0.0 |
| --wavelength <m> | X-ray wavelength in meter; default: 0.1e-9 | |
| --axis | Do not simulate but write axis for radial averaging with IM2DAT | |
| --mask | Do not simulate but write a centered detector mask for radial averaging with IM2DAT | |
| --mask-offset-x <X> | Mask offset in horizontal direction; default: 0.0 | |
| --mask-offset-y <Y> | Mask offset in vertical direction; default: 0.0 | |
| --header | Do not simulate but write header information with IM2DAT | |
| -o | --output <FILE> | Output file name (default: basename of the input file, extension depends on other options) |
| --format <FORMAT> | Output format, one of: .tiff, .edf, .cbf, .msk, … (default depends on other options) | |
| -v | --version | Print version information and exit. |
| -h | --help | Print a summary of arguments, options, and exit. |
Note that a mask offset of (0.0, 0.0) corresponds to an incident beam at the center of the simulated detector image; a mask-offset-y of half the detector height would correspond to an incident beam centered at the top edge of the detector.
Runtime Output
There is no output of IMSIM at run-time. For data output see output files.
imsim Input Files
IMSIM requires a calculated SAS intensity data (.abs) file. The angular grid of the input has to be sufficiently dense.
For proteins, .abs files may be obtained from CRYSOL, and for geometric shapes from BODIES.
imsim Output Files
IMSIM writes multiple output files, depening on the options provided:
| Option | Extension | Description |
|---|---|---|
| --axis | .dat | Generates an angular axis file that is suitable for the use with the --axis option of IM2DAT. The default output filename is the basename of the input data with extension .dat. Use the output option for a user-defined output file name and extension. |
| --mask | .msk | Generates a beamstop and detector mask definition for the use with the --beamstop-mask option of IM2DAT](im2dat.html#command-line-arguments-and-options). The default output filename is the basename of the input data with extension .msk. Use the output option for a user-defined output file name and extension. |
| --header | Generates a key-value file for the use as optional argument for IM2DAT. The default output filename is the basename of the input data with extension .txt. Use the output option for a user-defined output file name and extension. | |
| .tiff | Generates an image file for the use as argument for IM2DAT. The default output filename is the basename of the input data with extension .tiff. Use the output option for a user-defined output file name and extension. |
Please note that existing files with the same file name will be overwritten without further notice.
Examples
To simulate scattering data based on an available atomic model, first run CRYSOL, then use the resulting .abs file as an input for the simulation (sim.sh : helper script for bash- shells on Linux, Mac and Linux Subsystem for Windows; the setup corresponds roughly to the 3m position at EMBL BioSAXS beamline P12 in Hamburg, Germany). Note that the maximum scattering vector (option sm/smax in CRYSOL) must be sufficiently large to allow for the whole data range to be mapped to the detector. Here, for 1V18 :
$ crysol --smax 1.0 --ns 5001 --lm 50 1V18.pdb
$ bash sim.sh 1V1800.abs
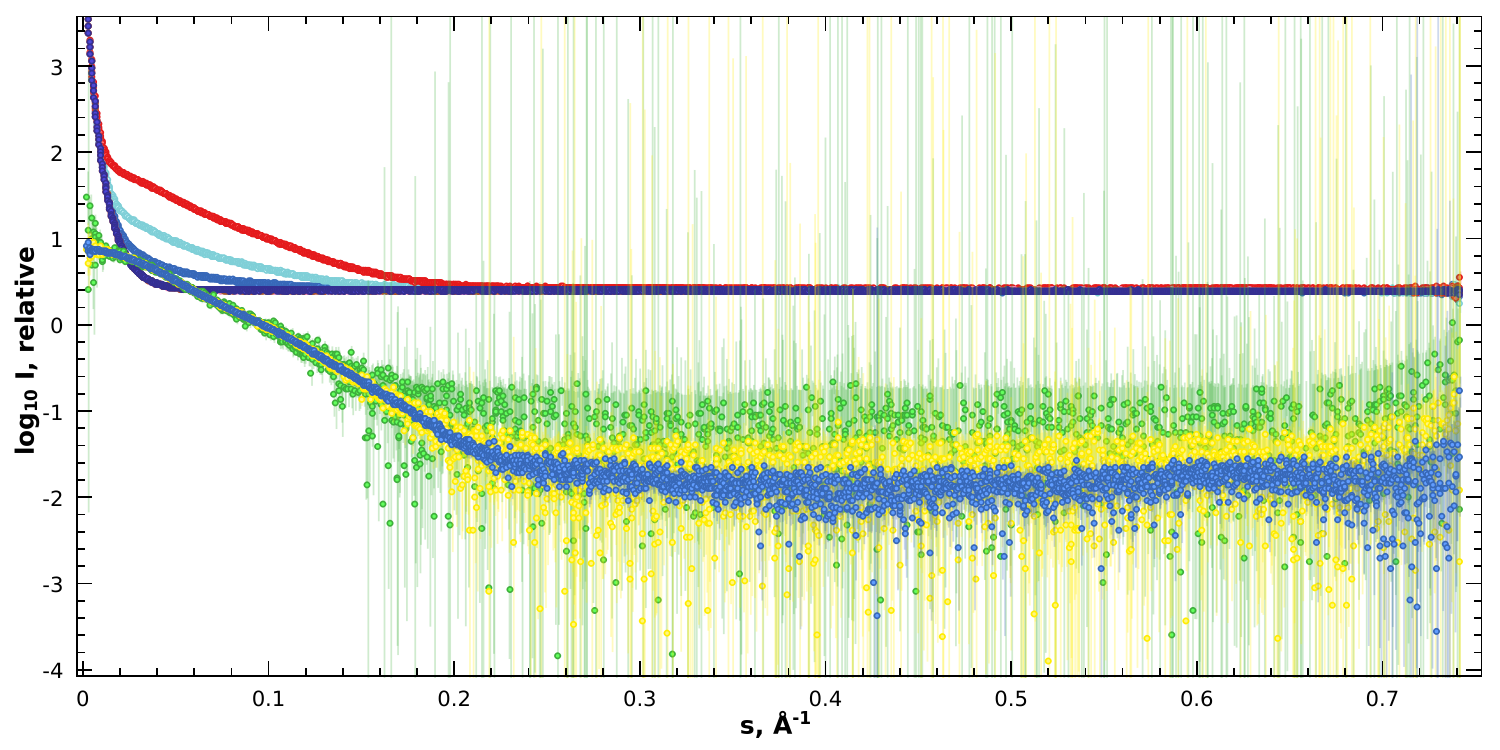
The results as shown in PRIMUS; a plot of simulated data
files of sample and background on logarithmic scale. In addition their
corresponding concentration-normalized subtractions for c=0.5,
c=2.0 and c=8.0 are displayed.
For Windows command interpreter there is a similar helper script, sim.bat. Its usage is:
> sim.bat <STRUCTURE_FILE> <CONCENTRATION>
Where STRUCTURE_FILE is the name of a *.pdb or *.cif file and CONCENTRATION is the value in mg/ml.